Publications
Structure and Dynamics dictate the Functional Destiny of Genomic DNA across Multiple Organisms [PUBLISHED]
DOI: https://doi.org/10.1016/j.ijbiomac.2025.147488
Download PDF: https://www.sciencedirect.com/science/article/pii/S0141813025080456?dgcid=coauthor
Abstract: DNA is a dynamic molecule composed of numerous genic and regulatory elements that orchestrate cellular functions. Traditional methods often fail to provide accurate functional genome annotations because they do not effectively account for sequence variability within and across different organisms. To address this, we conducted an extensive genomic physical fingerprinting of ∼4.6 million genomic elements. Utilizing multi-microsecond molecular dynamics simulation-derived parameters over higher nucleotide steps, we uncovered critical details about the physicochemical profiles of key DNA elements, including Coding Sequences, Promoters, Gene boundaries, Exon-Intron boundaries, Start Codons, and Stop Codons across the four diverse eukaryotic kingdoms. This exploration marks a significant advancement in functional genomics and opens new avenues for genome annotation, enhancing our understanding of eukaryotic genome complexity. We identified characteristic biophysical signals associated with key genomic sites across all 11 organisms studied. Our results demonstrate that closely related organisms exhibit similar patterns, underscoring the universality of our features. Furthermore, an investigation in human Enhancers and 5′/3’ UTR boundaries support this novel structural and energetic framework to annotate genomic elements with unprecedented accuracy. Lastly, sequence to structure and dynamics to function was an established axiom in proteomics. Our results demonstrate that this is true for nucleic acids as well.
Exon-Intron Boundary Detection Made Easy by Physicochemical Properties of DNA [PUBLISHED]
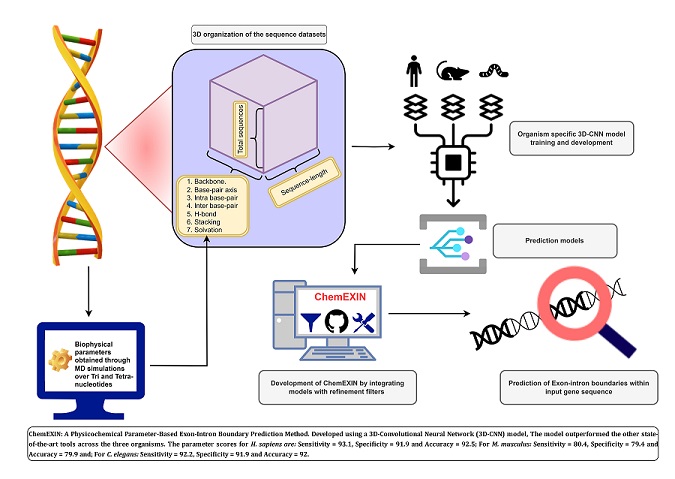
DOI: https://doi.org/10.1039/D4MO00241E
Download PDF: Click here Related Poster: Click here
Preprint: https://doi.org/10.21203/rs.3.rs-4359229/v1
Abstract: Genome architecture in eukaryotes exhibits a high degree of complexity. Amidst the numerous intricacies, the existence of genes as non-continuous stretches composed of exons and introns has garnered significant attention and curiosity among researchers. Accurate identification of exon–intron (EI) boundaries is crucial to decipher the molecular biology governing gene expression and regulation. This includes understanding both normal and aberrant splicing, with aberrant splicing referring to the abnormal processing of pre-mRNA that leads to improper inclusion or exclusion of exons or introns. Such splicing events can result in dysfunctional or non-functional proteins, which are often associated with various diseases. The currently employed frameworks for genomic signals, which aim to identify exons and introns within a genomic segment, need to be revised primarily due to the lack of a robust consensus sequence and the limitations posed by the training on available experimental datasets. To tackle these challenges and capitalize on the understanding that DNA exhibits function-dependent local physicochemical variations, we present ChemEXIN, an innovative novel method for predicting EI boundaries. The method utilizes a deep-learning (DL) architecture alongside tri- and tetra-nucleotidebased structural and energy features. ChemEXIN outperforms existing methods with notable accuracy and precision. It achieves an accuracy of 92.5% for humans, 79.9% for mice, and 92.0% for worms, along with precision values of 92.0%, 79.6%, and 91.8% for the same organisms, respectively. These results represent a significant advancement in EI boundary annotations, with potential implications for understanding gene expression, regulation, and cellular functions.
Book Chapters
Deep learning in computer-aided drug design: a case study [PUBLISHED]
DOI: https://doi.org/10.1016/B978-0-443-22299-3.00012-8
Download PDF: Click here
Abstract: Computer-aided drug design (CADD) is a rapidly growing field that combines the knowledge of computational chemistry, bioinformatics, and pharmacology to aid in the discovery and development of new drugs. CADD utilizes various computational techniques and tools such as molecular modeling, docking, and machine learning (ML) to predict the properties and interactions of potential drug compounds with their biological targets. One of the significant advantages of CADD is its ability to significantly reduce the time and cost of drug development by allowing for the virtual screening of many compounds in silico instead of relying solely on experimental methods, which can also reduce the number of compounds that must be tested in animal models and clinical trials. As a result, CADD has become an essential tool in modern pharmaceutical research. Molecular dynamics (MD) simulations, which may foretell the motion and interactions of atoms and molecules in a biological system, and docking, which can foretell the binding of a medicinal compound to its target protein, are two examples of CADD techniques. Artificial neural networks (ANNs) and random forests are two ML techniques used in CADD for various tasks, including virtual screening, lead optimization, absorption, distribution, metabolism, and excretion (ADMET) prediction.
The Role of Artificial Intelligence and Machine Learning in Autoimmune Disorders [PUBLISHED]
DOI: https://doi.org/10.1007/978-981-99-9029-0_3
Download PDF: Click here
Abstract: The immune system of an organism responds to threats from outside the body. Autoimmunity in immunology is the system of an organism's immune responses against its healthy cells, tissues, and other typical body parts. Therefore, a disorder known as an autoimmune illness results from an inappropriate immune reaction to a healthy bodily function. The science of developing computers with intelligence that both mimics and exceeds that of humans is known as artificial intelligence (AI). Programs having AI capabilities can contextualize and analyze data to deliver information or automatically initiate operations without the need for human intervention. Furthermore, AI can be attained through machine learning. This branch of AI applies to learning to make ever-better judgments using algorithms to discover patterns and acquire insights from data automatically. In this chapter, we present the role of AI and machine learning to analyze how autoimmunity behaves in conditions like autoimmune diseases. Moreover, we also discussed the impact of employing machine learning techniques to optimize precision medicine for patients.